This is an automated email from the ASF dual-hosted git repository.
thomasdelteil pushed a commit to branch master
in repository https://gitbox.apache.org/repos/asf/incubator-mxnet.git
The following commit(s) were added to refs/heads/master by this push:
new b8b352d Cleaned up profiling tutorial (#15228)
b8b352d is described below
commit b8b352d55bafed2910e5771d33982dea78989454
Author: Thom Lane <[email protected]>
AuthorDate: Thu Jun 13 13:47:58 2019 -0700
Cleaned up profiling tutorial (#15228)
* Cleaned up profiling tutorial.
* Minor edits to profiler tutorial.
* fix ascii issue
* Updates based on feedback.
---
docs/tutorials/python/profiler.md | 112 ++++++++++++-----------
docs/tutorials/python/profiler_nvprof.png | Bin 235747 -> 0 bytes
docs/tutorials/python/profiler_nvprof_zoomed.png | Bin 254663 -> 0 bytes
docs/tutorials/python/profiler_winograd.png | Bin 75450 -> 0 bytes
4 files changed, 59 insertions(+), 53 deletions(-)
diff --git a/docs/tutorials/python/profiler.md
b/docs/tutorials/python/profiler.md
index 8080309..9eed452 100644
--- a/docs/tutorials/python/profiler.md
+++ b/docs/tutorials/python/profiler.md
@@ -17,11 +17,11 @@
# Profiling MXNet Models
-It is often helpful to understand what operations take how much time while
running a model. This helps optimize the model to run faster. In this tutorial,
we will learn how to profile MXNet models to measure their running time and
memory consumption using the MXNet profiler.
+It is often helpful to check the execution time of each operation in a neural
network. You can then determine where to focus your effort to speed up model
training or inference. In this tutorial, we will learn how to profile MXNet
models to measure their running time and memory consumption using the MXNet
profiler.
## The incorrect way to profile
-If you have just begun using MXNet, you might be tempted to measure the
execution time of your model using Python's `time` module like shown below:
+If you have just started to use MXNet, you might be tempted to measure the
execution time of your model using Python's `time` module like shown below:
```python
from time import time
@@ -34,35 +34,19 @@ y = nd.dot(x, x)
print('Time for matrix multiplication: %f sec\n' % (time() - start))
start = time()
-print(y.asnumpy())
-print('Time for printing the output: %f sec' % (time() - start))
+y_np = y.asnumpy()
+print('Time for converting to numpy: %f sec' % (time() - start))
```
-
**Time for matrix multiplication: 0.005051 sec**<!--notebook-skip-line-->
-[[501.1584 508.29724 495.65237 ... 492.84705 492.69092 490.0481
]<!--notebook-skip-line-->
-
- [508.81058 507.1822 495.1743 ... 503.10526 497.29315
493.67917]<!--notebook-skip-line-->
-
- [489.56598 499.47015 490.17722 ... 490.99945 488.05008
483.28836]<!--notebook-skip-line-->
-
- ...<!--notebook-skip-line-->
-
- [484.0019 495.7179 479.92142 ... 493.69952 478.89194 487.2074
]<!--notebook-skip-line-->
-
- [499.64932 507.65094 497.5938 ... 493.0474 500.74512
495.82712]<!--notebook-skip-line-->
-
- [516.0143 519.1715 506.354 ... 510.08878 496.35608
495.42523]]<!--notebook-skip-line-->
+**Time for converting to numpy: 0.167693 sec**<!--notebook-skip-line-->
-**Time for printing the output: 0.167693 sec**<!--notebook-skip-line-->
+From the timings above, it seems as if converting to numpy takes lot more time
than multiplying two large matrices. That doesn't seem right.
+This is because, in MXNet, all operations are executed asynchronously. So,
when `nd.dot(x, x)` returns, the matrix multiplication is not complete, it has
only been queued for execution. However,
[`asnumpy`](http://mxnet.incubator.apache.org/api/python/ndarray/ndarray.html?highlight=asnumpy#mxnet.ndarray.NDArray.asnumpy)
has to wait for the result to be calculated in order to convert it to numpy
array on CPU, hence taking a longer time. Other examples of 'blocking'
operations include [`as [...]
-From the output above, it seems as if printing the output takes lot more time
that multiplying two large matrices. That doesn't feel right.
-
-This is because, in MXNet, all operations are executed asynchronously. So,
when `nd.dot(x, x)` returns, the matrix multiplication is not complete, it has
only been queued for execution. `asnumpy` in `print(y.asnumpy())` however,
waits for the result to be computed and hence takes longer time.
-
-While it is possible to use `NDArray.waitall()` before and after operations to
get running time of operations, it is not a scalable method to measure running
time of multiple sets of operations, especially in a Sequential or Hybrid
network.
+While it is possible to use
[`NDArray.waitall()`](http://mxnet.incubator.apache.org/api/python/ndarray/ndarray.html?highlight=waitall#mxnet.ndarray.waitall)
before and after operations to get running time of operations, it is not a
scalable method to measure running time of multiple sets of operations,
especially in a
[`Sequential`](http://mxnet.incubator.apache.org/api/python/gluon/gluon.html?highlight=sequential#mxnet.gluon.nn.Sequential)
or hybridized network.
## The correct way to profile
@@ -70,7 +54,10 @@ The correct way to measure running time of MXNet models is
to use MXNet profiler
```python
from mxnet import profiler
-profiler.set_config(profile_all=True, aggregate_stats=True,
filename='profile_output.json')
+
+profiler.set_config(profile_all=True,
+ aggregate_stats=True,
+ filename='profile_output.json')
```
`profile_all` enables all types of profiling. You can also individually enable
the following types of profiling:
@@ -84,10 +71,11 @@ profiler.set_config(profile_all=True, aggregate_stats=True,
filename='profile_ou
### Setup: Build a model
-Let's build a small convolutional neural network that we can use for profiling.
+Let's build a small convolutional neural network that we can use to
demonstrate profiling.
```python
from mxnet import gluon
+
net = gluon.nn.HybridSequential()
with net.name_scope():
net.add(gluon.nn.Conv2D(channels=20, kernel_size=5, activation='relu'))
@@ -103,11 +91,13 @@ We need data that we can run through the network for
profiling. We'll use the MN
```python
from mxnet.gluon.data.vision import transforms
-train_data =
gluon.data.DataLoader(gluon.data.vision.MNIST(train=True).transform_first(transforms.ToTensor()),
- batch_size=64, shuffle=True)
+
+dataset = gluon.data.vision.MNIST(train=True)
+dataset = dataset.transform_first(transforms.ToTensor())
+dataloader = gluon.data.DataLoader(dataset, batch_size=64, shuffle=True)
```
-Let's define a method that will run one training iteration given data and
label.
+Let's define a function that will run a single training iteration given `data`
and `label`.
```python
# Use GPU if available
@@ -120,37 +110,33 @@ else:
net.collect_params().initialize(mx.init.Xavier(), ctx=ctx)
# Use SGD optimizer
-trainer = gluon.Trainer(net.collect_params(), 'sgd', {'learning_rate': .1})
+trainer = gluon.Trainer(net.collect_params(), 'sgd', {'learning_rate': 0.1})
-# Softmax Cross Entropy is a frequently used loss function for multi-classs
classification
+# Softmax Cross Entropy is a frequently used loss function for multi-class
classification
softmax_cross_entropy = gluon.loss.SoftmaxCrossEntropyLoss()
# A helper function to run one training iteration
def run_training_iteration(data, label):
-
# Load data and label is the right context
data = data.as_in_context(ctx)
label = label.as_in_context(ctx)
-
# Run the forward pass
with autograd.record():
output = net(data)
loss = softmax_cross_entropy(output, label)
-
# Run the backward pass
loss.backward()
-
# Apply changes to parameters
trainer.step(data.shape[0])
```
### Starting and stopping the profiler from Python
-When the first forward pass is run on a network, MXNet does a number of
housekeeping tasks including inferring the shapes of various parameters,
allocating memory for intermediate and final outputs, etc. For these reasons,
profiling the first iteration doesn't provide accurate results. We will,
therefore skip the first iteration.
+When the first forward pass is run on a network, MXNet does a number of
housekeeping tasks including inferring the shapes of various parameters,
allocating memory for intermediate and final outputs, etc. For these reasons,
profiling the first iteration doesn't provide representative results for the
rest of training. We will, therefore, skip the first iteration.
```python
# Run the first iteration without profiling
-itr = iter(train_data)
+itr = iter(dataloader)
run_training_iteration(*next(itr))
```
@@ -164,18 +150,21 @@ profiler.set_state('run')
run_training_iteration(*next(itr))
-# Ask the profiler to stop recording after operations have completed
+# Make sure all operations have completed
mx.nd.waitall()
+# Ask the profiler to stop recording
profiler.set_state('stop')
```
Between running and stopping the profiler, you can also pause and resume the
profiler using `profiler.pause()` and `profiler.resume()` respectively to
profile only parts of the code you want to profile.
-### Starting profiler automatically using environment variable
+### Starting the profiler automatically using an environment variable
The method described above requires code changes to start and stop the
profiler. You can also start the profiler automatically and profile the entire
code without any code changes using the `MXNET_PROFILER_AUTOSTART` environment
variable.
-MXNet will start the profiler automatically if you run your code with the
environment variable `MXNET_PROFILER_AUTOSTART` set to `1`. The profiler output
is stored into `profile.json` in the current directory.
+`$ MXNET_PROFILER_AUTOSTART=1 python my_script.py`
+
+MXNet will start the profiler automatically if you run your code with the
environment variable `MXNET_PROFILER_AUTOSTART` set to `1`. The profiler output
is stored in `profile.json` inside the current directory.
Note that the profiler output could be large depending on your code. It might
be helpful to profile only sections of your code using the `set_state` API
described in the previous section.
@@ -183,9 +172,11 @@ Note that the profiler output could be large depending on
your code. It might be
MXNet executes computation graphs in 'bulk mode' which reduces kernel launch
gaps in between symbolic operators for faster execution. This could reduce the
granularity of the profiler output. If you need profiling result of every
operator, please set the environment variables `MXNET_EXEC_BULK_EXEC_INFERENCE`
and `MXNET_EXEC_BULK_EXEC_TRAIN` to `0` to disable the bulk execution mode.
+When working with networks created using the Gluon API, you will get a more
granular profiling outputs if you profile networks that haven't been
hybridized. Operations can appear fused together in the profiling outputs after
hybridization, which can make debugging tricky.
+
### Viewing profiler output
-There are a few ways to view the information collected by the profiler. You
can view it in the console, you can view a more graphical version in a browser,
or you can use a vendor tool such as Intel VTune or Nvidia NVProf to view
output. For most scenarios the information you need can be obtained with
MXNet's built in profiler support, but if you want to investigate the
performance of operators along side extra context about your hardware (e.g.
cache hit rates, or CUDA kernel timings) th [...]
+There are a few ways to view the information collected by the profiler. You
can view it in the console, you can view a more graphical version in a browser,
or you can use a vendor tool such as Intel VTune or Nvidia NVProf to view
output. For most scenarios the information you need can be obtained with
MXNet's built in profiler support, but if you want to investigate the
performance of operators alongside extra context about your hardware (e.g.
cache hit rates, or CUDA kernel timings) the [...]
#### 1. View in console
@@ -215,29 +206,44 @@ Let's zoom in to check the time taken by operators
The above picture visualizes the sequence in which the operators were executed
and the time taken by each operator.
-#### 3. View in NVProf
+## Advanced: Using NVIDIA Profiling Tools
-You can view all MXNet profiler information alongside CUDA kernel information
by using the MXNet profiler along with NVProf. Use the MXNet profiler as in
the samples above, but invoke your python script with the following wrapper
process available on most systems that support CUDA:
+MXNet's Profiler is the recommended starting point for profiling MXNet code,
but NVIDIA also provides a couple of tools for low-level profiling of CUDA
code:
[NVProf](https://devblogs.nvidia.com/cuda-pro-tip-nvprof-your-handy-universal-gpu-profiler/),
[Visual Profiler](https://developer.nvidia.com/nvidia-visual-profiler) and
[Nsight Compute](https://developer.nvidia.com/nsight-compute). You can use
these tools to profile all kinds of executables, so they can be used for
profiling Python [...]
-```bash
-nvprof -o my_profile.nvvp python my_profiler_script.py
-==11588== NVPROF is profiling process 11588, command: python
my_profiler_script.py
-==11588== Generated result file:
/home/kellen/Development/incubator-mxnet/ci/my_profile.nvvp
-```
-Your my_profile.nvvp file will automatically be annotated with NVTX ranges
displayed alongside your standard NVProf timeline. This can be very useful
when you're trying to find patterns between operators run by MXNet, and their
associated CUDA kernel calls.
+#### NVProf and Visual Profiler
+
+NVProf and Visual Profiler are available in CUDA 9 and CUDA 10 toolkits. You
can get a timeline view of CUDA kernel executions, and also analyse the
profiling results to get automated recommendations. It is useful for profiling
end-to-end training but the interface can sometimes become slow and
unresponsive.
+
+You can initiate the profiling directly from inside Visual Profiler or from
the command line with `nvprof` which wraps the execution of your Python script.
If it's not on your path already, you can find `nvprof` inside your CUDA
directory. See [this discussion
post](https://discuss.mxnet.io/t/using-nvidia-profiling-tools-visual-profiler-and-nsight-compute/)
for more details on setup.
+
+`$ nvprof -o my_profile.nvvp python my_profiler_script.py`
+
+`==11588== NVPROF is profiling process 11588, command: python
my_profiler_script.py`
-
+`==11588== Generated result file:
/home/user/Development/incubator-mxnet/ci/my_profile.nvvp`
-In this picture we see a rough overlay of a few types of information plotted
on a horizontal timeline. At the top of the plot we have CPU tasks such as
driver operations, memory copy calls, MXNet engine operator invocations, and
imperative MXNet API calls. Below we see the kernels active on the GPU during
the same time period.
+We specified an output file called `my_profile.nvvp` and this will be
annotated with NVTX ranges (for MXNet operations) that will be displayed
alongside the standard NVProf timeline. This can be very useful when you're
trying to find patterns between operators run by MXNet, and their associated
CUDA kernel calls.
-
+You can open this file in Visual Profiler to visualize the results.
+
+
+
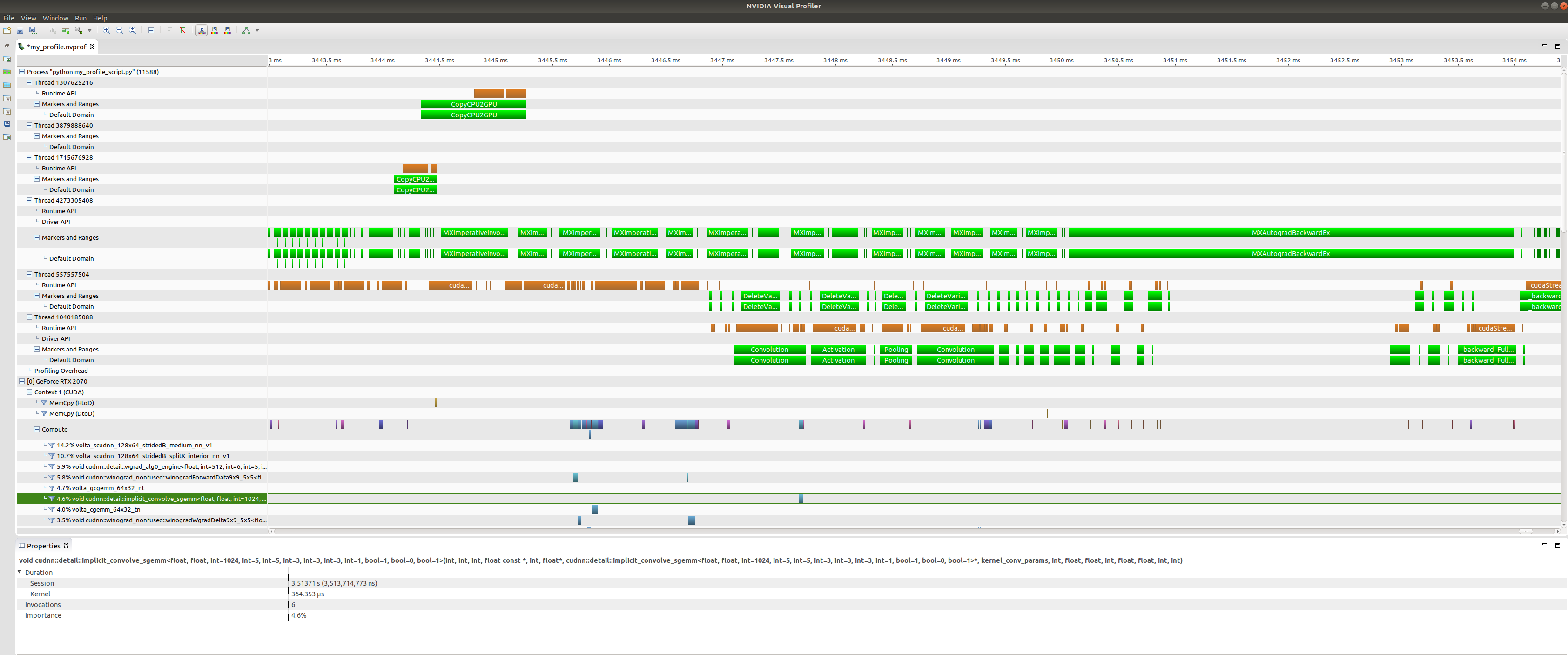
+At the top of the plot we have CPU tasks such as driver operations, memory
copy calls, MXNet engine operator invocations, and imperative MXNet API calls.
Below we see the kernels active on the GPU during the same time period.
+
+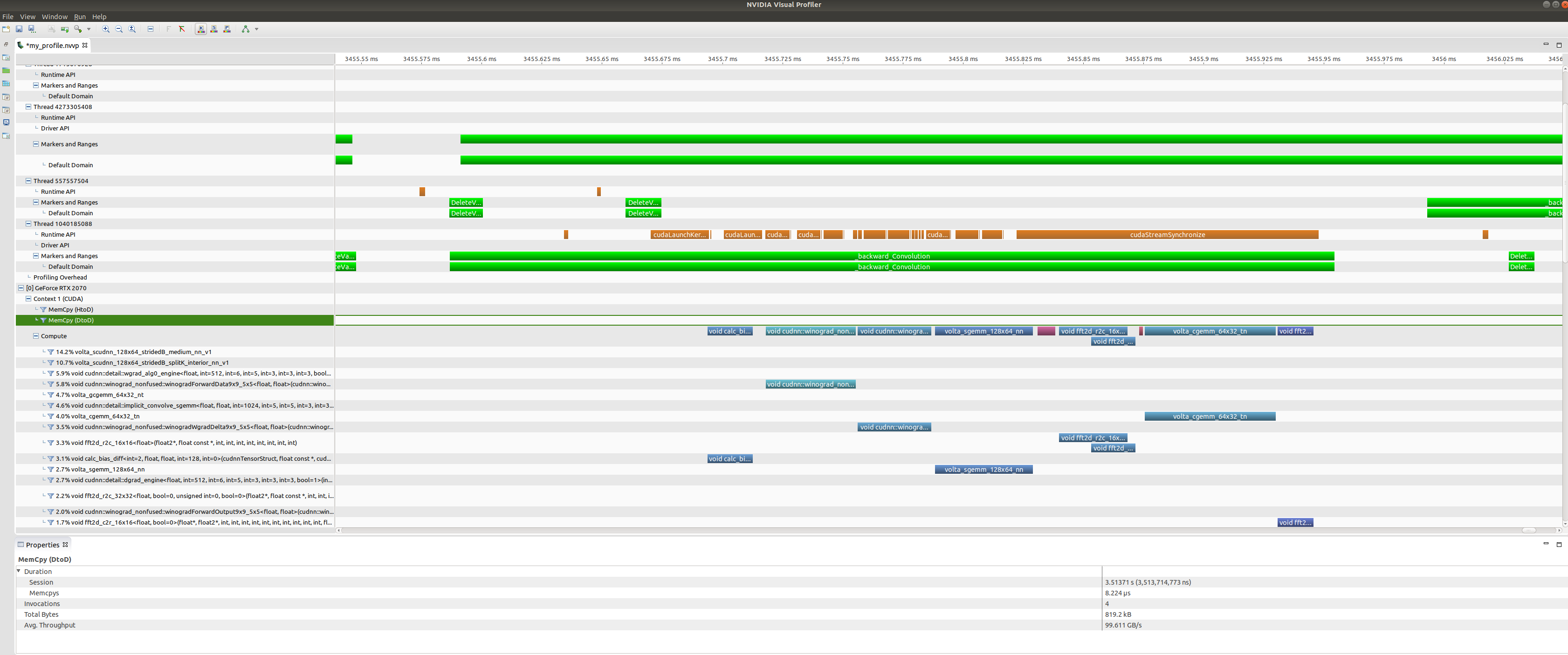
Zooming in on a backwards convolution operator we can see that it is in fact
made up of a number of different GPU kernel calls, including a cuDNN winograd
convolution call, and a fast-fourier transform call.
-
+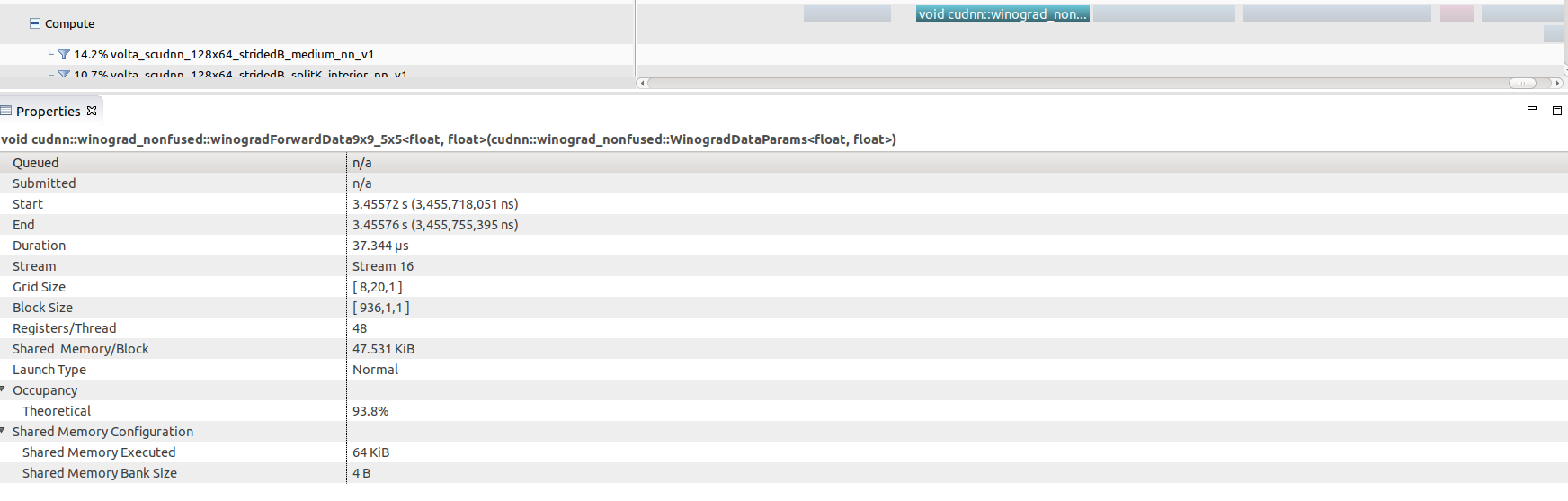
Selecting any of these kernel calls (the winograd convolution call shown here)
will get you some interesting GPU performance information such as occupancy
rates (vs theoretical), shared memory usage and execution duration.
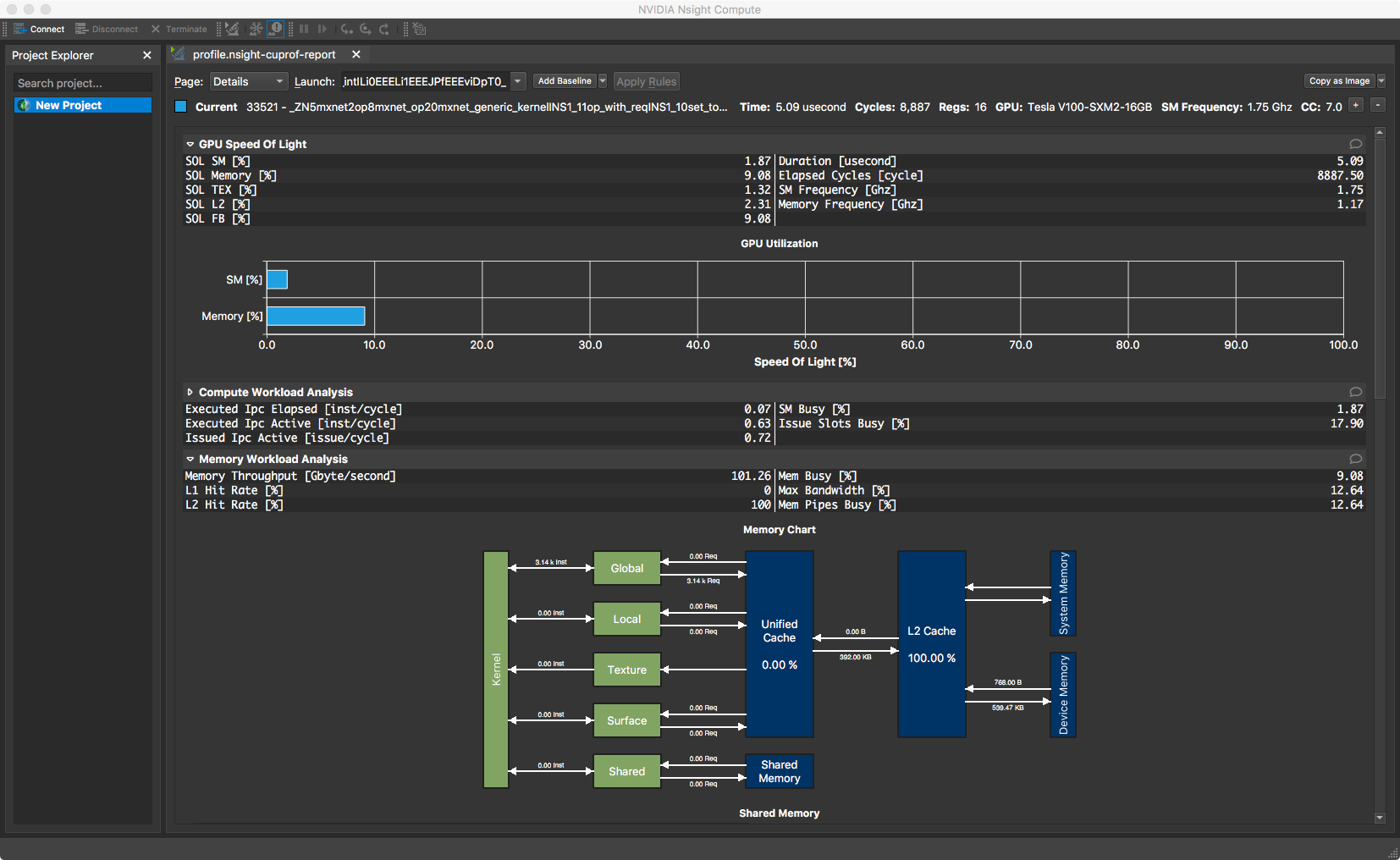
+#### Nsight Compute
+
+Nsight Compute is available in CUDA 10 toolkit, but can be used to profile
code running CUDA 9. You don't get a timeline view, but you get many low level
statistics about each individual kernel executed and can compare multiple runs
(i.e. create a baseline).
+
+
+
### Further reading
- [Examples using MXNet
profiler.](https://github.com/apache/incubator-mxnet/tree/master/example/profiler)
diff --git a/docs/tutorials/python/profiler_nvprof.png
b/docs/tutorials/python/profiler_nvprof.png
deleted file mode 100644
index 37d8615..0000000
Binary files a/docs/tutorials/python/profiler_nvprof.png and /dev/null differ
diff --git a/docs/tutorials/python/profiler_nvprof_zoomed.png
b/docs/tutorials/python/profiler_nvprof_zoomed.png
deleted file mode 100644
index 9b6b6e8..0000000
Binary files a/docs/tutorials/python/profiler_nvprof_zoomed.png and /dev/null
differ
diff --git a/docs/tutorials/python/profiler_winograd.png
b/docs/tutorials/python/profiler_winograd.png
deleted file mode 100644
index 5b4fcc3..0000000
Binary files a/docs/tutorials/python/profiler_winograd.png and /dev/null differ
