qsourav opened a new issue, #14629:
URL: https://github.com/apache/arrow/issues/14629
Hi,
It seems like the compute method, filter() has some performance issue, when
the number of True values are very small in the input mask in comparison to the
number of false values. It is even slower than pandas dataframe filter
operation.
Although filter() is slower, but the take() method seems to have better
performance for those cases.
Please consider the following experiemental code:
---
import time
import math
import numpy as np
import pyarrow as pa
parr = pa.array(range(6161464))
def get_pos(N, per):
tot = math.ceil(N * per)
step = N // tot
ret = []
for i in range(0, N, step):
ret.append(i)
return ret
def eval(N):
pers = [i * 0.003 for i in range(1, 30)]
for i in pers:
per = round(i * 100, 2)
indx = get_pos(N, i)
stime = time.time()
ret = parr.take(indx)
print("[per: {}%] pyarrow take time: {} sec".format(per,
time.time() - stime))
mask = np.asarray([False] * N)
mask[indx] = True
pa_mask = pa.array(mask)
stime = time.time()
ret = parr.filter(pa_mask)
print("[per: {}%] pyarrow filter time: {} sec".format(per,
time.time() - stime))
eval(6161464)
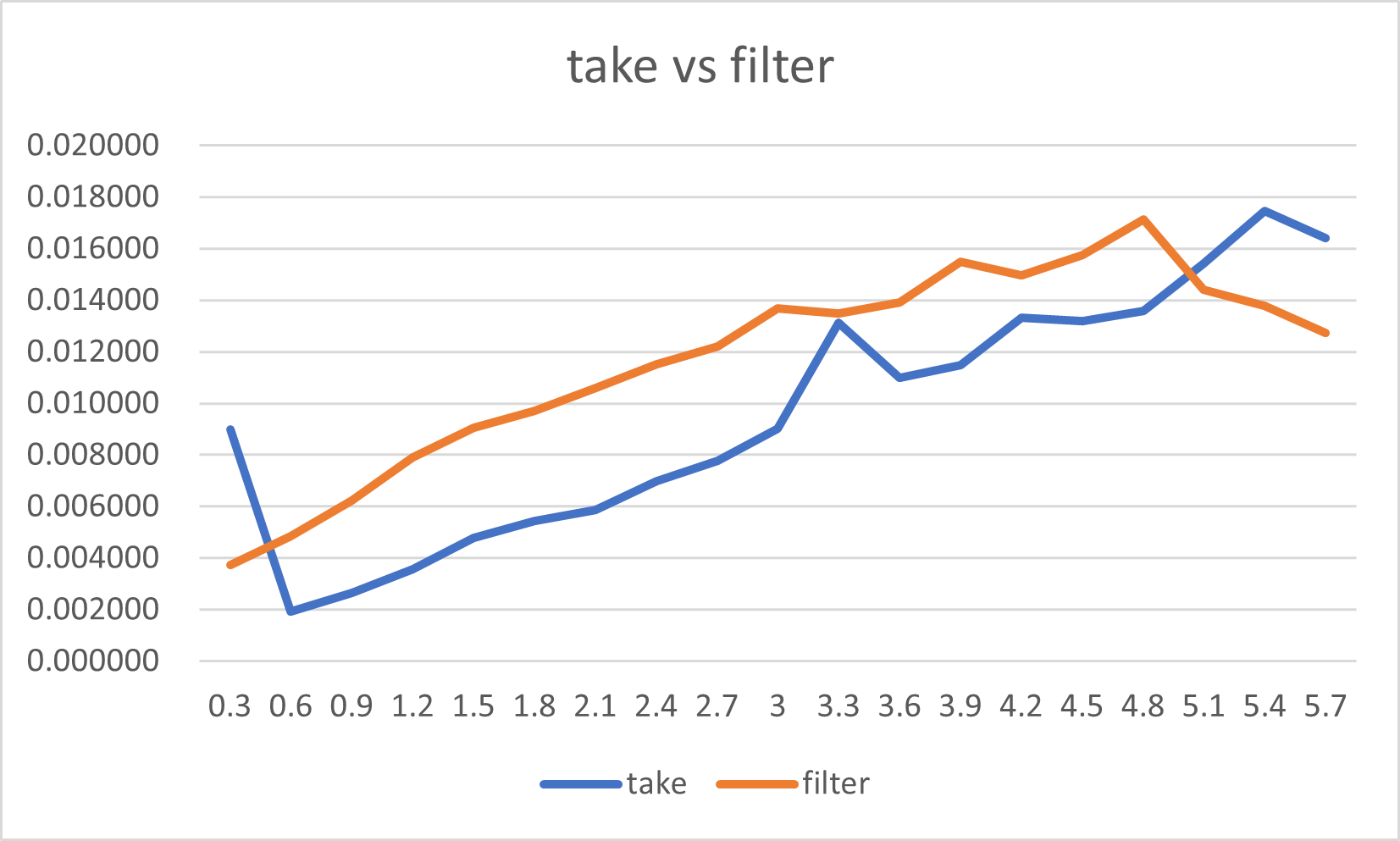
The take() method is significantly faster for the distribution of True
values less than 5.1% in the input mask.
Filter starts showing better performance when the "True"-distribution
exceeds 5.1%.
Filter is a very frequently used method and having lesser True values is
somewhat very common in data preprocessing.
Thus it would be really great if the perfiormance issue of Filter() can be
checked and improved.
Thanks,
Sourav
--
This is an automated message from the Apache Git Service.
To respond to the message, please log on to GitHub and use the
URL above to go to the specific comment.
To unsubscribe, e-mail: [email protected]
For queries about this service, please contact Infrastructure at:
[email protected]
